|
Nat Genet (2023) [website]
Mouse embryonic development requires transposable element expression ————————- “The paper was a joy to read, the figures were presented clearly, it was well-referenced, and I think it will be of great interest to both the TE community and early embryo developmental biologists. Resolving the true function of MERVL in development and ZGA is an important goal for the field.” Dr. Edward J. Grow, UT Southwestern Medical Center, Dallas, TX, USA. |
||
|
Nat Cell Biol (2021) [website]
Golden opportunity for piRNA in female fertility Yongjuan Guan & P. Jeremy Wang ————————- Notably, three studies of golden hamsters by Hasuwa et al.10, Loubalova et al.11 and Zhang et al.12 in this issue of Nature Cell Biology demonstrate that PIWI proteins, MOV10L1 and piRNAs are essential for the production of both functional oocytes and male gametes in mammals. |
||
|
Trends in Genetics, Available online 6 October (2019) [PDF]
De novo DNA Methylation: Who’s Your DADdy? Marla E.Tharp, Alex Bortvin ————————- DNA methylation regulates the organization and function of the genome. Yamanaka et al. now report that de novo methylation of male germ cells of mice involves the transient opening of heterochromatin at megabase-size differentially accessible domains (DADs). This chromatin remodeling likely facilitates de novo methylation of the germ cell genome. |
||
|
Nature Structural and Molecular Biology 26, 758–765 (2019) [PDF]
An RNA exporter that enforces a no-export policy David Homolka and Ramesh S. Pillai ————————- Co-transcriptional repression can be viewed as a dynamic hierarchical process ultimately resulting in deposition of silent histone marks. Murano et al. examined Nxf2-tethered-reporter silencing in a time-resolved manner to arrive at the conclusion that silencing is a two-step process. At early time points, reporter silencing is achieved through reduced RNA polymerase II occupancy, without deposition of H3K9me3 marks. Only long-term silencing is correlated with the presence of silent histone marks. |
||
|
Cell 167, 310–312 (2016) [PDF]
PIWI Takes a Giant Step Yibel Xiao and Ailong Ke ————————- In this issue of Cell, Matsumoto et al. (2016) unveil the crystal structure of the silkworm Siwi-piRISC complex. The success doesn’t come easy, as it involves raising a monoclonal antibody to affinity purify Siwi-piRISC directly from BmN4 cells and using a protease to release the PIWI region for structure determination. The payoff, however, is well worth the effort. The authors note structural differences of Siwi with the AGO-clade relatives, reveal piRNA recognition features, and examine the slicing mechanism. These observations lay a solid foundation for more sophisticated mechanistic dissections in the future. |
||
|
Nature Reviews Molecular Cell Biology 14, 544–545 (2013) [PDF]
PIWI’s new assistant Rachel David ————————- Previous work had shown that GTSF1 is required for transposon silencing in mouse testes. Muerdter et al., Ohtani et al. and Dönertas et al. knocked down the homologue of GTSF1 in D. melanogaster and found that this blocked silencing of piRNA-targeted transposons. Interestingly, the phenotype of D. melanogaster lacking GTSF1 was comparable to those of Piwi–piRNA pathway mutants. These findings suggest a key role for GTSF1 in transposon silencing as part of the Piwi–piRNA pathway. |
||
|
Science vol. 339: 25-27 (2013) [PDF]
The Immune System’s Compact Genomic Counterpart Mitch Leslie ————————- Other animals bolster their piRNAs directly, relying on what’s called the pingpong amplifi cation loop. Hannon’s team and a group led by molecular biologist Mikiko Siomi, now at Keio University School of Medicine in Tokyo, independently described this mechanism in flies in 2007, but mice and zebrafish also take advantage of a similar process. In the ping-pong loop, piRNAs and Piwi proteins slice up transposon RNA. The resulting fragments undergo modifi cation and join with other Piwi proteins to cut up RNA transcripts of piRNA clusters, thus making new piRNAs (see diagram, p. 26). The loop “only amplifi es the useful piRNAs,” those with a target available in the cell, says Brennecke, a co-author on one of the papers. |
||
|
Nature Structural & Molecular Biology vol. 19: 11 (2012) [PDF]
Zucchini has bite Arianne Heinrichs ————————- The protein Zucchini belongs to the phospho- lipase D (PLD) family of phos-phodiesterases and has been implicated in promoting primary PIWI-interacting RNA (piRNA) biogenesis. PLD family members include both phospho-lipases and nucleases, but it has been unclear whether Zucchini has an impact on the piRNA biogenesis pathway indirectly (through a putative phospholipase activity) or directly (as a putative nuclease). Three recent reports address this question by analyzing mouse and Drosophila melanogaster Zucchini (mZuc and DmZuc, respectively) using structural and functional approaches. Ipsaro et al. determined the crystal structure of N-terminally truncated mZuc, whereas Nishimasu et al. and Voigt et al. solved the structure of an equivalent truncation of DmZuc. The papers demonstrate that Zucchini forms a dimer, with the active site assembled from conserved residues of both monomers, consistent with known monomeric and dimeric PLD protein structures. The mZuc structure contains an unexpected CCCH- type zinc-finger motif, which has been implicated in the binding of single-stranded RNA molecules, whereas DmZuc has a CHCC-type zinc-finger motif. In addition, the active site forms a narrow groove that can accommodate single-stranded, but not double-stranded, nucleic acids. Ipsaro et al. modeled a short RNA molecule into the structure of mZuc, illustrating the shape and charge complementarity provided by the RNA substrate. In support of the structural data, in vitro analyses confirmed that mZuc and DmZuc lacked any detectable phospholipase activity but instead demonstrated single-strand–specific nuclease activity. Nishimasu et al. further validated the functional significance of DmZuc’s nuclease activity by demonstrating that wild-type dimeric DmZuc is critical for transposon silencing. mZuc and DmZuc cleavage generates 5′ phosphate and 3′ hydroxyl termini, which suggests that Zucchini might generate the 5′ ends of mature primary piRNAs, which are known to bear a 5′-monophosphate group. Future studies will be needed to pinpoint Zucchini’s precise role in primary piRNA biogenesis. |
||
|
Nature Reviews Genetics, AOP, published online 22 Nov. (2011) [PDF] RESEARCH HIGHLIGHTS RNAi gets stuck into transcription Mary Muers ————————- RNAi is best known for its various roles in post-transcriptional gene silencing in the cytoplasm. It is also known to have nuclear functions associated with repressive chromatin or gene silencing, such as the formation of heterochromatin in some species. Researchers have now uncovered a rather different nuclear role for some key RNAi proteins: interaction with the core transcriptional machinery at transcriptionally active loci. Through a series of cellular and chromatin fractionation experiments in Drosophila melanogaster cells, Cernilogar et al. showed that the RNAi components Dicer 2 (DCR2) and Argonaute 2 (AGO2) are mainly nuclear and associated with chromatin. In vivo staining with antibodies against these proteins revealed that they localize to euchromatic, transcriptionally active regions of D. melanogaster polytene chromosomes, including loci containing copies of the heat-shock gene Hsp70. The authors found that depletion of AGO2 or DCR2 resulted in increased expression of heat-shock genes under non-heat-shock conditions. A key mode of transcriptional regulation of the heat-shock genes is RNA polymerase II (RNAPII) pausing; prior to heat shock, RNAPII stops a short distance downstream of the transcriptional start site and heat shock triggers transcriptional elongation. Through further experiments, including chromatin immunoprecipitation to look at the distribution of RNAPII, Cernilogar et al. showed that these RNAi proteins are involved in maintaining RNAPII pausing. This influence on pausing also occurs at loci other than heat-shock genes: in particular, AGO2 and DCR2 seem to be involved in the global transcriptional repression of non-heat-shock genes that occurs after heat shock, and their influence on RNAPII dynamics requires their enzymatic activity. The authors examined the relationship between these RNAi proteins and the core transcriptional machinery in more detail. They found that DCR2 and AGO2 interact with RNAPII and negative elongation factor E (NELFE) and that they also influence the association of RNAPII with NELFE; this might be a mechanism by which the RNAi proteins influence RNAPII dynamics. However, small RNAs also seem to be involved in the activity of the RNAi proteins at active loci: high-throughput sequencing of RNAs that are associated with AGO2 produced small RNA tags that map across the transcription unit of active genes. This distribution suggests a possible influence of AGO2 on RNAPII processivity as well as pausing. The AGO2associated RNAs were derived from the antisense strand, and so they might target AGO2 to sense transcripts. This work reveals a novel sphere of influence of RNAi. It will be interesting to explore whether this involvement in RNAPII activity occurs in other species and also to further dissect the mechanisms and the types of genes that it acts on. |
||
|
Development vol. 138, 3093 (2011) [PDF]
RNA silencing in Monterey Olivia S. Rissland and Eric C. Lai ————————- Kuniaki Saito, from the laboratory of Mikiko and Haru Siomi (Keio University, Japan), described a complementary Bm-N4 cellbased assay for secondary piRNA production. Saito reported that immunopurified Siwi cleaved a complementary target, yielding an expected 47-nt product that reflected the generation of a secondary piRNA precursor. Strikingly, when performed in lysates, this cleavage reaction produced a 28-nt product, which was interpreted as a mature secondary piRNA generated by piRISC cleavage and subsequent trimming by unknown factors. With these revelations of in vitro piRNA biogenesis, the hunt is undoubtedly on to establish analogous systems from more genetically tractable species, as well as to purify the nucleases and other factors that mediate piRNA biogenesis. |
||
|
The New York Academy of Sciences [WEB] NYAS eBriefing on “piRNA” meeting Biogenesis and Function of Piwi-Interacting RNAs (piRNAs) ————————- Another researcher, Haruhiko Siomi, from Keio University School of Medicine, studies how transposable elements are silenced in germline cells. “Recently we and other people found Piwi and piRNAs are involved in transposon silencing but we still don’t know how they are involved in the silencing,” he said. By studying the role of piRNAs in Drosophila ovaries, he has found that piRNAs are produced in the follicle (somatic) cells of the ovary in addition to the germ cells. To better understand what proteins are important for biogenesis of piRNAs in ovarian somatic cells, he used an RNAi technique to identify genes that, when knocked down, result in the loss of piRNAs in these cells. This uncovered three proteins, including Armitage, Yb, and Zucchini that were required for piRNA biogenesis. Furthermore, loss of these proteins in ovarian somatic cells led to increased levels of transposon transcripts, the target of many piRNAs. Siomi showed that the Armitage and Yb proteins interact and co-localize at the Yb bodies in the cell cytoplasm. Interestingly, Armitage binds to a number of intermediate forms of piRNA in addition to mature piRNAs, suggesting a model by which these piRNA intermediates first interact with Armitage within the Yb body to be processed into mature piRNAs that then associate with Piwi proteins before moving into the nucleus to silence their target RNAs. |
||
|
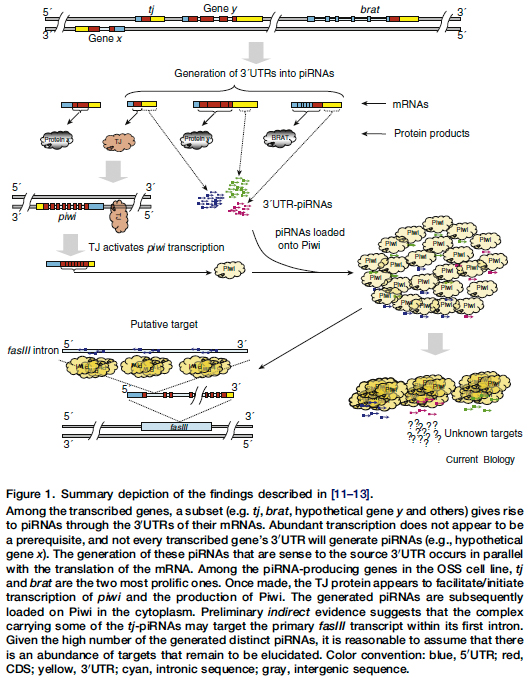
Science, Vol. 327, 394 (2010) [PDF] EDITORS’ CHOICE Untranslated Regulators A variety of short noncoding RNA molecules―microRNAs, small interfering RNAs, and Piwiinteracting RNAs (piRNAs)―play regulatory roles in eukaryotes. Many piRNAs are derived from transposon-related sequences and, through complementary sequence interactions and a “ping-pong” amplification process, act to silence those selfish and potentially mutagenic elements in germline cells. However, Robine et al. show that a substantial population of piRNAs found in a Drosophila somatic ovarian cell line are in fact derived from a distinct subset of genes, and also that the bulk of these piRNAs arise directly from the 3’ untranslated regions (3’ UTRs) of the sense strands. This suggests that the complementary targets of these piRNAs may not be the parental transcripts. Ping-pong amplification is not required for the generation of these 3’ UTR piRNAs, nor does it appear as if they are aberrant products of the primary piRNA processing pathway. Furthermore, Saito et al. have found that the Drosophila gene traffic jam (tj) gives rise to 3’ UTR piRNAs and that one of its targets is the fasciclin III gene transcript, and Robine et al. note that the subset of functional categories of mRNAs that gives rise to the 3’ UTR piRNAs is broadly conserved between fruit flies and mice. ― GR Curr. Biol. 19, 2066 (2009); Nature 461, 1296 (2009). |
||
|
Genome Biology, Vol. 11, 304 (2010) [PDF] MEETING REPORT mRNA: a complex(ed) life Michaela Muller, Karla M Neugebauer and Christian Eckmann A report of the EMBL conference ‘The Complex Life of mRNA: From Synthesis to Decay’, Heidelberg, Germany, 18-21 March 2010. ————————- In contrast, germline-enriched PIWI proteins are known to contain symmetrically dimethylated arginine. Mikiko Siomi (Keio University, Tokyo, Japan) has discovered that arginine methylation enhances Ago protein complex formation and the efficient loading of small RNAs in Drosophila. Interestingly, Siomi reported that tudor-domain proteins are sensitive to arginine-modified Ago proteins and may therefore represent key regulators of piRNA-directed silencing complexes. Thus, signaling cascades may regulate post-transcriptional controls during development or in response to changing environmental conditions. |
||
|
Silence, Vol. 1, 8 (2010) [PDF] REVIEW Riding in silence: a little snowboarding, a lot of small RNAs Stefan L Ameres, Ryuya Fukunaga
Meeting report of the recent symposium, RNA silencing: Mechanism, Biology and Applications, organized by Phillip D. Zamore (University of Massachusetts Medical School) and Beverly Davidson (University of Iowa), and held in Keystone, Colorado. ————————- Mikiko Siomi (Keio University School of Medicine) described the specific association of a Tudor domain-containing protein, Tudor, with Aubergine and Ago3 through symmetric dimethyl-arginine (sDMA) modifications in the PIWI proteins introduced by dPRMT5/Capsuleen/DART5. She suggested that this might be involved in the loading of primary PIWI-interacting (pi) RNAs into the effector PIWI proteins [28]. Studies in a fly ovarian somatic cell line expressing PIWI but not Aubergine or Ago3, indicate that PIWI and the RNA helicase Armitage (Armi) but not Maelstrom (Mael) or Spindle-E (Spn-E) (both factors implicated in the piRNA pathway in flies) are required for piRNA accumulation. Armi co-localizes with the helicase and Tudor domaincontaining protein, Yb, to distinct foci in the cytoplasm called Yb bodies. Similar colocalization can be observed in fly ovaries and testes. In a model, PIWI and Armi were proposed to be involved in primary piRNA accumulation in Yb bodies, whereas Mael and Spn-E might participate in the ‘ping-pong’ amplification loop. |
||
|
RNA Biology, Vol. 7, (2010) [PubMed] POINT OF VIEW Transposon defense in Drosophila somatic cells A model for distinction of self and non-self in the genomed Klaus Forstemann A New Splice Variant of Loquacious is Required for Endo-siRNA Generationz When they were initially discovered, it came as a big surprise that endo-siRNAs depend on the double-stranded RNA binding domain (dsRBD) protein Loquacious (Loqs) rather than its homolog R2D2, which is required for the function of exogenous siRNAs (1-4). In particular, it was puzzling how Loqs could help to recognize pre-miRNAs This manuscript has been published online, prior to printing. Once the issue is complete and page numbers have been assigned, the citation will change accordingly. with imperfect base-pairing in concert with Dcr-1 and at the same time endosiRNA precursors with perfect base pairing together with Dcr-2. Three publications now report the identification of a previously unkown variant of Loqs that is dedicated to endo-siRNA processing (5,6,7) This variant preferentially interacts with Dcr-2, rather than Dcr-1, and thus establishes endo-siRNA biogenesis as a pathway with specific components, rather than a mix-and-match between the known miRNA and siRNA pathways. Perhaps most importantly, this provides a possibility to specifically impair only endo-siRNA generation while leaving the other pathways intact. 7.Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA, 2010
|
||
|
Current Biology, Vol. 20, R110 [PDF] Dispatch Short RNAs: How Big Is This Iceberg? Isidore Rigoutsoss
Recent studies have reported the identification of piwi-associated RNAs (piRNAs) in Drosophila somatic cells. Interestingly, these piRNAs derive from the 30 untranslated regions of a subset of transcribed protein-coding genes and, experimentation suggests, might control the expression of other protein-coding transcripts. Studies of additional organisms support the new pathway’s presence across animals. |
||
 |
||
|
Nature Cell Biology, Vol. 11, 1285 [PDF] RESEARCH HIGHLIGHTS Traffic jam in the piRNA world Germline-specific PIWI-interacting RNAs (piRNAs) are produced either through a primary pathway, which loads them onto the argonaute protein Piwi, or through an amplification loop, which involves additional activities of the Piwi-related proteins Aubergine and AGO3. Whereas the amplification loop is fairly well understood, far less is known about the primary production of piRNAs. Siomi and colleagues uncover a new locus, traffic jam (TJ), which produces piRNA in the somatic lineage of gonads through this primary route (Nature, doi: 10.1038/nature08501). The authors derived an ovarian somatic cell line (OSC) from Drosophila that expressed Piwi, but neither Aubergine nor AGO3. They found that in OSC cells, Piwi associated with piRNAs that were mostly antisense to retrotransposons. Several piRNAs in OSC cells do not exhibit the characteristics of amplification loop products shown by those derived from the known flamenco piRNA locus and 3′UTR of the gene encoding the transcription factor TJ. Somatic cells mutant for TJ or Piwi fail to envelop germline stem cells, and the adhesion molecule Fasciclin III, known to be involved in this process, is upregulated in these cells. Piwi is absent from TJ-mutant gonadal somatic cells and TJ can bind the Piwi promoter. Thus TJ regulates primary piRNA production in the somatic lineage by acting on Piwi, possibly at the transcriptional level, and by providing a source of piRNAs. In turn, the Piwi pathway can regulate gonadal features, perhaps through piRNA-mediated control of adhesion molecules. NLB
|
||
| Nature Review Genetics, Vol. 10, 738 [PDF]RESEARCH HIGHLIGHTSSmall RNAsA regulatory circuit for piwi by the large Maf gene traffic jam in DrosophilaSaito, K. et al. Nature 7 Oct 2009 (10.1038/nature08501)Piwi-interacting RNAs (piRNAs) silence retrotransposons in Drosophila germ lines by binding to Piwi proteins (Piwi, Argonaute 3 and Aubergine), but the primary processing of piRNAs has been elusive. Using a Drosophila ovarian somatic cell line, these authors identified a new locus, traffic jam (tj), that encodes piRNAs, and they define a novel regulatory circuit that simultaneously produces two different molecules from tj mRNA: the transcription factor TJ, which activates Piwi expression, and piRNAs that direct Piwi to its target genes. | ||
| Current Biology, Vol. 18, R561-563 [PDF]DispatchRNA Interference: Endogenous siRNAs Derived from Transposable ElementsDarren J. Obbard1 and David J. FinneganSummaryThe Piwi-interacting RNA interference pathway plays an important role in suppressing transposable elements in the Drosophila germline. Now, deep sequencing of short RNAs from somatic tissue and cell culture has identified a novel class of endogenous siRNAs that may have a similar role in the soma.————————-In a further twist, Kawamura et al.[14] observe signs of RNA editing in their siRNAs, and this can also be seen in the data of Chung et al.[11], reinforcing the idea that RNA editing and RNAi pathways may interact [20]. | ||
| Nature Structural & Molecular Biology, 15, 546 (2008) [PDF]NEWS AND VIEWSEndo-siRNAs: yet another layer of complexity in RNA silencingTimothy W Nilsen————————-To address this question, Czech et al.2 and Kawamura et al.3 used immunoprecipitation of AGO2 in the hope of identifying endogenous small RNAs associated with it. Both groups obtained strikingly similar results. Each identified populations of small RNAs that are clearly distinct from previously characterized piRNAs or miRNAs, and both found that most of these small RNAs, dubbed esiRNAs, were derived from mobile genetic elements. Other esiRNAs were processed from either hairpin structures or from overlapping RNAs formed by convergent transcription. | |
| Cell, 133, 747-748 (2008) [PDF]Leading EdgeMolecular Biology Select————————-Argonaute proteins are essential components of the RNAi machinery that associate with distinct classes of small RNAs to exert their effector functions. One branch of the Argonaute family, the PIWI subfamily of proteins, form complexes with Piwi-interacting RNAs (piRNAs) and are essential for restricting the activity of transposons in the germline. Argonaute proteins are associated with small interfering RNAs (siRNAs) or microRNAs (miRNAs), and silence gene expression by either siRNA guided cleavage of the target mRNA transcript, or by miRNA-mediated posttranscriptional repression involving both translational inhibition and/or mRNA degradation. In Drosophila there are three PIWI proteins and two proteins of the Argonaute family, AGO1 and AGO2. Genetic and biochemical evidence has demonstrated functional specialization in fly AGO proteins, with AGO1 binding to miRNAs and AGO2 being associated with siRNA-mediated gene silencing. Functional specialization extends to the biogenesis pathways associated with these small RNAs; miRNAs are processed from endogenous hairpin precursors by cleavage events involving the RNaseIII enzymesDrosha and Dicer1 (Dcr-1) with its partner Loquacious (Loqs). siRNAs loaded into AGO2 are processed from long dsRNAs by Dicer2 (Dcr-2) and its partner R2D2, but until recently only siRNAs from exogenous long dsRNAs had been reported in flies and mammals. A flurry of recent papers (Ghildiyal et al., 2008; Czech et al., 2008; Kawamura et al., 2008; Okamura et al., 2008) now report the isolation of endogenous siRNAs (endo-siRNAs) from both somatic and gonadal cells of Drosophila and provide insight into their biological functions. Interestingly, endo-siRNAs appear to be important in both genome protection and gene regulation, thus bridging roles normally associated with piRNAs and miRNAs. Also, by combining factors involved in the biogenesis of both exogenous siRNAs and miRNAs, they add complexity to our current view of the biogenesis pathways of small RNAs in Drosophila. | |
| NateureReviews, 9, (2008) [PDF]Endo-siRNAs truly endogenous————————-.Endo-siRNAs were also reported in D. melanogaster gonadal and somatic cells by the groups of Greg Hannon, Eric Lai, Haruhiko and Mikiko Siomi, and Phil Zamore. Many endo-siRNAs correspond to transposons, heterochromatic sequences, and cis-natural antisense transcripts, and can target both protein-coding genes and mobile elements. | |||
| Science, 320, 1023-1024 (2008) [PDF]Slicing and Dicing for small RNAJames A. Birchler and Harsh H. KaviA new type of small RNA and mode of gene regulation is discovered in fly and mammals.————————-In a complementary study, Kawamura et al. (6) sequenced small RNAs bound by Ago2 in cultured Drosophila cells. The endosiRNAs recovered showed homology to a subset of transposons. Depletion of Dicer-2 and Ago2 in these cells reduced the quantities of endo-siRNAs and increased the expression of the corresponding transposons. Endo-siRNAs with homology to transposons were also found in fly ovaries, and their function depended on Dicer-2 and Ago2, revealing that the germ line has both siRNA- and piRNA-generating machineries. | |
| EMBO reporst, 8, 723-729 (2007) [PDF]meeting reportDelving into the diversity of silencing pathwaysSymposiumm on MicroRNAs and siRNAs: Biological functions and MechanismsSilke Dorner, Ana Eulalio, Eric Huntzinger & Elisa Izaurralde————————-Ping-pong mechanism for rasiRNA biogenesisPIWI-like proteins are required for the establishment and maintenance of the germline. In Drosophila, these proteins are thought to have a role in silencing retrotransposons and other repetitive genetic elements, thereby preserving the integrity of the genome. Last year, several groups reported that small RNA s associated with these proteins do not have the characteristic 21-nucleotide length of miRNA s or siRNA s, but are 25-30 nucleotides in length (reviewed by Parker & Barford, 2006). Furthermore, the biogenesis of these RNA s is independent of Dicer (Vagin et al, 2006), raising the question of how they are generated.
G. Hannon (Cold Spring Harbor, NY , USA) and M. Siomi (Tokushima, Japan) have provided at least a partial answer to this question (Brennecke et al, 2007; Gunawardane et al, 2007). Both groups have analysed small RNA s associated with the Drosophila PI WI- like protein Argonaute 3 (AGO 3). Similarly to the two other Drosophila PI WI- like proteins-PI WI and Aubergine (AU B)-AGO 3 is expressed mainly in the germline, and associates with small 24-27 nucleotide RNA s that are complementary to AU B-associated small RNA s in their first ten nucleotides. The PI WI- and AU B-associated RNA s match the antisense strand of retrotransposons and repetitive sequence elements (therefore referred to as rasiRNA s), whereas AGO 3-associated RNA s are derived from the sense strand. PI WI- and AU B-associated RNA s show a strong bias for uracil (U) at their 5′ ends, whereas AGO 3-associated RNA s show a strong preference for adenine (A) at position 10. Together, these observations suggest an amplification loop mechanism whereby the 5′ end of AU B- (or PI WI)- associated RNA s are generated by endonucleolytic cleavage of precursor transcripts guided by AGO 3-associated RNA s. Conversely, the 5′ ends of AGO 3-associated RNA s are generated by endonucleolytic cleavage guided by rasiRNA s associated with AU B (or PI WI; Fig 1). In agreement with this model, Siomi and co-workers have shown that both AU B and AGO 3 have slicing activity.
|
|||
| Genes & Development, 21, 1707-1713 (2007) [PubMed]REVIEWpiRNAs-the ancient hunters of genome invadersJulia Verena Hartig, Yukihide Tomari, and Klaus Forstemann——A Drosophila homolog of HEN1, called Pimet/DmHen1, appends the 2′-O-Me modification onto piRNAs (Horwich et al. 2007; Saito et al. 2007). pimet/hen1 encodes a single-strand-specific methyltransferase that lacks the dsRNA-binding domain of plant HEN1; piRNAs isolated from pimet/hen1 mutant ovaries are unmodified. Recombinant Pimet/DmHen1 protein can bind to Aub, Piwi, and Ago3 complexes isolated from pimet/hen1 mutant ovaries and methylate the piRNAs within Aub complexes. This suggests that the 3′-end resection of piRNA precursors precedes methylation. In contrast, Pimet/DmHen1 does not bind to Ago1 or methylate miRNA/miRNA* duplexes or single-stranded miRNAs bound to Ago1. | |||
| Mol Cell, 26, 603 (2007) [PDF]The Coming of Age for Piwi ProteinsAnita G. Seto, Robert E. Kingston and Nelson C. LauPiwi proteins, a subfamily of Argonaute (Ago) proteins, have recently been shown to bind endogenous small RNAs. However, differences between Ago proteins (which bind microRNAs and small interfering RNAs) and Piwi proteins and Piwi-interacting RNAs (piRNAs) suggest novel functions for Piwi proteins. Here, we highlight the recent progress in understanding Piwi function and the implications for germline and stem cell development. —————In 2006, six groups converged upon the landmark finding that Piwi proteins in flies and mammals bind a class of small RNAs distinct from miRNAs and typical siRNAs. Two groups (Saito et al., 2006; Vagin et al., 2006) determined that Piwi and Aubergine bind repeat-associated siRNAs (rasiRNAs). The rasiRNAs were first characterized in small RNA cloning studies from flies at different stages of development, which revealed that the rasiRNAs were longer in length than canonical small RNAs (24-27 nucleotides, as opposed to 21-22), were enriched in the testes and early in development, and derived from retrotransposons and other repetitive elements (Aravin et al., 2003). In addition to their longer length, rasiRNAs can be distinguished from miRNAs by a modification to the 30 terminal nucleotide (Vagin et al., 2006). rasiRNAs copurify with Piwi, but not with DmAgo1, which binds miRNAs. Consistent with the Piwi protein and the rasiRNAs working together, an endonuclease activity on a target RNA complementary to a short guide RNA in the protein could be detected (Gunawardane et al., 2007; Saito et al., 2006). | |||
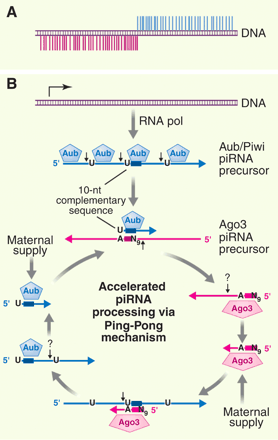
| Science, 316, 397 (2007) [PDF]ReviewpiRNAs in the Germ LineHaifan LinSmall noncoding RNAs have emerged as potent regulators of gene expression at both transcriptional and posttranscriptional levels. Recently, a class of small RNAs that interact with Piwi proteins has been discovered in the mammalian germ line and Drosophila. These Piwi-interacting RNAs (piRNAs) represent a distinct small RNA pathway. | ||||||
 |
||||||
—————In Drosophila, some piRNAs have been called repeat-associated siRNAs (“rasiRNAs”) (3, 4, 6). However, unlike siRNAs, these rasiRNAs bind to Piwi, Aubergine (Aub), and Ago3ムthree Piwi subfamily proteins, but not to Ago subfamily proteins. Moreover, their production requires neither Dicer-1 nor Dicer-2, which generate miRNAs and siRNAs, respectively (3). Thus, by definition, these rasiRNAs are piRNAs.How are piRNAs produced? They are likely produced from long single-stranded precursors by yet-to-be-identified endonucleases. In Drosophila, Piwi, Aub, and Ago3 could be such endonucleases because they have slicing activity (4, 6). For some transposon-derived piRNAs in Drosophila, an additional “Ping-Pong” mechanism might be involved in accelerating their processing from precursors (5, 6) (Fig. 1B). This is possible because the Aub- and Piwi-associated piRNAs match the antisense strand of DNA, showing a strong bias for a uracyl (U) at the 5′ ends; yet Ago3-associated piRNAs match the sense strand and complement Aub- and Piwi-associated piRNAs in their first 10 nucleotides, showing a conserved adenine (A) at position 10. Thus, the Ago3-piRNA complex might guide and cleave the 5′ ends of Aub- and Piwi-associated piRNAs, whereas the Aub-Piwi-piRNA complex might guide and cleave the 5′ ends of Ago3-associated piRNAs (Fig. 1B).(ref. 6) L.S. Gunawardane et al. |
||||||
| Fig. 1. (A) A bidirectional piRNA cluster. (B) A proposed piRNA biogenesis pathway. The resulting piRNA-protein complexes then regulate gene expression at the epigenetic or posttranscriptional levels (not shown). N9, the 9th nucleotide from A10 in Ago3-piRNA; the question mark (?), other nuclease. | ||||||
| Nature, 446, 864 (2007) [PDF]NEWS & VIEWSGenomic defence with a slice of piPhillip D. ZamoreIn fruit flies, a few very large genes generate the small RNAs that silence parasitic DNA elements. These RNAs might also participate in an amplification circuit that increases their potency.—————The Hannon and Siomi laboratories separately set out to identify small RNAs bound to each of the three fly Piwi proteins, in the hope that their sequences would reveal how rasiRNAs are made and function. Their findings are simply spectacular, suggesting that rasiRNAs arise from a small number of trigger loci – huge ‘genes’ that produce small RNAs against many selfish genetic elements – and that they are amplified through reciprocal cycles of cleavage by pairs of Piwi proteins. | |||
| Development, 134, 1635 (2007) [PDF]Meeting ReviewThe regulation of genes and genomes by small RNAsVictor Ambros and Xuemei ChenRecent advances have illuminated the surprisingly diverse modes of small RNA-guided gene regulation by Argonaute-based mechanisms (Fig. 1). The Piwi class of Argonaute proteins is essential for spermatogenesis in mice and for gametogenesis in flies (Cox et al., 1998; Kim, 2006); in the past year or so, a new class of small RNAs ? piRNAs ? have been found to be associated with Piwi family proteins (Saito et al., 2006; Vagin et al., 2006; Grivna et al., 2006; Aravin et al., 2006; Girard et al., 2006; Lau et al., 2006). Although piRNA function in gametogenesis is still unclear, Gregory Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA) and Mikiko Siomi (University of Tokushima, Tokushima, Japan) reported progress in understanding the biogenesis and activity of Drosophilarepeat-associated siRNA (rasiRNAs), which are similar to mammalian piRNAs in many respects. | |||
| Cell, 129, 37 (2007) [PDF]Leading Edge ReviewMighty Piwis Defend the Germline against Genome IntrudersKathryn A. O’Donnell and Jef D. BoekePiwis are a germline-specific subclass of the Argonaute family of RNA interference (RNAi) effector proteins that are associated with a recently discovered group of small RNAs (piRNAs). Recent studies in Drosophila and zebrafish directly implicate Piwi proteins in piRNA biogenesis to maintain transposon silencing in the germline genome (Brennecke et al., 2007, Gunawardane et al., 2007, Houwing et al., 2007). This function may be conserved in mice as loss of Miwi2, a mouse Piwi homolog, leads to germline stem cell and meiotic defects correlated with increased transposon activity (Carmell et al., 2007). | |||
| Nature Structural & Molecular Biology, 13, 762 (2006) [PDF]Research highlightsPiwi pathway for retrotransposon silencing In the last few years, several pathways for RNA-mediated gene silencing have been defined in Drosophila melanogaster. The Argonaute (AGO) proteins AGO1 and AGO2, expressed ubiquitously throughout development, are essential in two of the pathways, with AGO1 binding micro-RNAs in one and AGO2 binding small interfering RNAs (siRNAs) in the other. Both activities lead to repression of transcripts complementary to the bound RNAs. The functions of other AGO-related proteins have remained relatively obscure, although mutants of Piwi, an AGO relative expressed primarily in germline stem cells, are known to affect the expression of retrotransposons. |
||||
| A new study by Siomi and colleagues further elucidates a silencing pathway involving specific interaction of Piwi with repeat-associated siRNAs (rasiRNAs) complementary to retrotransposons. The authors identified rasiRNAs bound to Piwi (but not to AGO1), whereas no miRNAs coimmunoprecipitated and siRNAs were not loaded onto Piwi in cell lysates. Unlike AGO1 and AGO2, Drosophila Piwi localizes in the nucleus; it also lacks the Asp-Asp-His catalytic motif shared by AGO1 and AGO2, yet has a similar ability to cleave target transcripts in vitro. The Piwi-bound rasiRNAs match transposable transcripts in both the sense and antisense directions. Chemical end groups on these RNAs imply that they are products of RNase III activity similar to that of the Dicer enzymes, which generate siRNAs and microRNAs and load them onto AGO proteins. As no such loading protein was found in Piwi immunoprecipitates, the factors and mechanisms that underlie rasiRNA processing remain to be determined. (Genes Dev. 20, 2214-2220, 2006) | ||||
 |
||||
| EurekAlert! AAAS Public release date: 31-Jul-2006 [URL]Identifying Piwi’s partnersPiwi-dependent gene silencingDr. Mikiko Siomi and colleagues from the Institute for Genome Rsesearch in Japan have identified the elusive small RNA partners of Piwi proteins lending new insight into the gene silencing pathways mediated by small RNAs in Drosophila.Piwi (P-element-induced wimpy testis) is a member of the Drosophila Argonaute protein family, which is expressed specifically in fly testis and ovaries, and is necessary for germ stem cell self-renewal. It has recently been shown that Piwi is responsible for the silencing of retrotransposons in the testis.Now, Dr. Siomi and colleagues demonstrate that in Drosophila ovaries, Piwi specifically associates with a distinct class of 25-29 nucleotide-long small RNAs, called repeat-associated siRNAs (rasiRNAs). They further show that Piwi is able to cleave target RNAs in vitro what the authors refer to as Slicer activity. According to the authors, their result suggest “that Piwi functions in nuclear RNA silencing as Slicer by associating specifically with the rasiRNAs,” thereby introducing a third, and novel, pathway of gene silencing in Drosophila. | |||
| Development, Vol. 133, 2451-2453 [PDF]Meeting ReviewNew inroads to developmentRalf J. Sommer——————————–The 2006 symposium of the RIKEN Center for Developmental Biology in Kobe, Japan, was entitled `Logic of development: new strategies and concepts’. The purpose of the meeting was to uncover how our understanding of the logic of developmental processes is changing in the light of novel techniques and strategies. The speakers provided a comprehensive overview of diverse topics, such as the power of functional genomics and imaging approaches, and the phenomenon of non-coding RNAs and their role in development. A wide range of processes and mechanisms at the molecular, cellular and organismic level were presented, as were many novel concepts and strategies, which together highlighted the importance of interdisciplinary approaches. ——————————–Haruhiko Siomi (University of Tokushima, Tokushima, Japan) demonstratedthe function of the Argonaute protein Ago2 in the DrosophilaRNAi pathway.
——————————– |
|||
| Developmental Cell, Vol. 10, 419-424 [PDF]Meeting ReviewDemystifying Small RNA PathwaysAmy E. Pasquinelli ——————————–The slicer function of some Argonaute proteins can play dual roles in the RNAi pathway. In addition to cleaving the target strand of an mRNA-siRNA duplex, the discarded passenger strand siRNA can also be sliced in half by Argonaute (Matranga et al., 2005, Miyoshi et al., 2005 and Rand et al., 2005). Mikiko Siomi (University of Tokushima, Japan) and Phillip Zamore (University of Massachusetts) each showed that Drosophila Ago2 can cleave the passenger strand of an siRNA duplex and that this event facilitates RISC maturation. It remains to be explained how cleavage of the passenger strand favors its displacement from Ago2.——————————– | |||
| Current Biology, Vol. 15, R603-605 [PDF]DispatchMicroRNAs: Loquacious Speaks outPhilipp J.F. Leuschner, Gregor Obernosterer and Javier Martinez In Drosophila, Dicer-2 requires the double-stranded RNA binding protein R2D2, to mediate the assembly of short interfering RNAs into the RNA-induced silencing complex. New data show that Dicer-1 also requires a double-stranded RNA binding protein called Loquacious for efficient microRNA-mediated gene silencing.A major breakthrough in molecular biology was the finding that eukaryotic cells harness mechanisms of RNA interference (RNAi) to regulate the expression of endogenous genes [1; 2]. Just as in the ‘canonical’ RNAi pathway, short single-stranded RNA molecules called microRNAs (miRNAs) serve as sequence specific guides to target silencing complexes containing an Argonaute (Ago) protein to cognate sequences typically in the 3′-untranslated region of target messenger RNAs [3].miRNAs originate from long primary transcripts (pri-miRNAs) [4], which are processed in the nucleus by the RNase III-like enzyme Drosha and its cofactor Pasha/DCGR8 into ~65 nt, hairpin shaped precursors, or pre-miRNAs [5; 6; 7; 8]. Pre-miRNAs are exported to the cytoplasm, where another RNase III-like enzyme, Dicer, liberates a ~22 nt long miRNA duplex from the hairpin. One strand of the duplex is integrated into an active RNA-induced silencing complex (RISC) [9].While human Dicer is able to process long double-stranded RNA (dsRNA) as well as pre-miRNAs, the two pathways are separated in Drosophila (Figure 1). Here, Dicer-2 does not play a role in miRNA biogenesis, but is required to cleave long dsRNAs into short interfering RNAs (siRNAs), which are then assembled into siRISCs (Figure 1A). Dicer-1 processes pre-miRNAs and loads the resulting miRNAs into miRISC containing Ago-1 (Figure 1B). However, Dicer-1 also seems to be required downstream of siRNA-production in siRISC assembly [10; 11; 12]. It has been shown that the activities of Drosha and Dicer-2 absolutely depend on the auxiliary dsRNA binding domain (dsRBD) proteins, Pasha and R2D2, respectively [5; 6; 7; 8; 13]. Now Saito et al. [14] and F?stemann et al. [15] provide a missing piece in the RNAi versus miRNA puzzle: they conclusively demonstrate that Dicer-1 also requires a dsRBD protein to efficiently process pre-miRNAs into miRNA duplexes. Saito et al. [14] relied on an RNAi-based functional screen for Drosophila dsRBD proteins that affect miRNA biogenesis, while F?stemann et al. [15] searched a database for conserved dsRBDs containing proteins. Both laboratories identified the same candidate ? a paralogue of Drosophila R2D2, featuring two canonical and one non-canonical dsRBDs. This candidate was baptized loquacious (loqs), as endogenous RNA-mediated silencing is lost in mutant flies. | |||||||
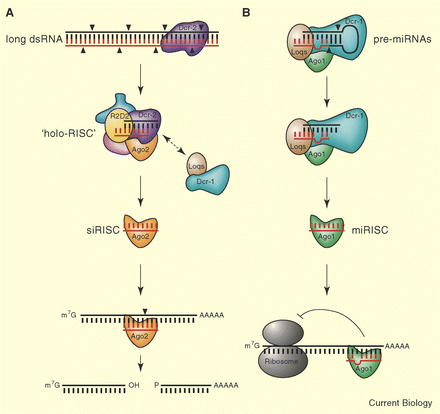 |
Figure 1. Model of the Dicer-1 and Dicer-2 pathways in Drosophila.(A) The RNase III-like enzyme Dicer-2 (Dcr-2) is needed to process long dsRNAs into duplex siRNAs. R2D2, a dsRBD protein, helps to sense duplex asymmetry and, together with Dicer-2, facilitates the assembly of an 80S ‘holo-RISC’ [16], a multi-protein complex. Both Dicer-1 and Loqs play a role downstream of siRNA production, probably as components of the putative ‘holo-RISC’ complex. Upon binding to complementary sequences in target RNAs, RISC cleaves target mRNAs. (B) A protein complex consisting of the RNase III-like enzyme Dicer-1 (Dcr-1), the dsRBD protein Loqs and Ago1 is able to convert pre-miRNA hairpins into ~22 nt miRNA duplexes, which are then loaded into the miRISC, composed of a single stranded miRNA bound to Ago1. This silencing complex inhibits translation by binding to complementary sequences in the 3′ untranslated region of cognate messenger RNAs. | ||||||
| Both groups show that reduced levels of Loqs result in the accumulation of endogenous pre-miRNAs. The same phenotype is observed when cells are depleted of Dicer-1, but not Dicer-2 or R2D2. The physical association of Loqs and Dicer-1 was confirmed by reciprocal co-immunoprecipitation, and did not depend on a pre-miRNA substrate. Saito et al. [14] demonstrate that both proteins also co-immunoprecipitate with Ago1, providing further evidence that miRNA processing may be directly linked to the assembly of miRISC. If assayed for processing activity in vitro, immunoprecipitates of Dicer-1 or Loqs readily generated mature miRNAs from synthetic precursors and were also found to associate with pre- and mature miRNAs in vivo.So what is the actual function of Loqs in this complex? It is certainly not simply the stabilization of Dicer-1, because Dicer-1 protein levels did not decrease significantly in the absence of Loqs. According to Saito et al. [14], Loqs confers substrate specificity for pre-miRNAs to Dicer-1. Surprisingly, Dicer-1 processes long dsRNA as well as pre-miRNA substrates, if Loqs is removed from the complex. Re-addition of Loqs inhibited dsRNA processing and enhanced pre-miRNA processing. But how is this achieved? Three splice variants of loqs are known, of which only two isoforms interact with Dicer-1. Interestingly, the third isoform lacks the non-canonical dsRBD, suggesting that this domain may be essential for association with Dicer-1. Loqs could enhance the binding efficiency of Dicer-1 to pre-miRNAs, thus stabilizing this interaction. Alternatively, it could position Dicer-1 in such a way that allows efficient processing, or introduce conformational changes in Dicer-1 leading to an enhancement of pre-miRNA cleavage.In order to assess the in vivo role of loqs in greater detail, F?stemann et al. [15] took advantage of a hypomorphic loqs mutant. These flies showed reduced silencing of a miRNA-controlled transgene as well as defects in silencing triggered by long dsRNA. Thus, Loqs is not only required for efficient miRNA-mediated silencing, but also cooperates with Dicer-1 in order to enhance silencing by siRNAs, emphasising the idea of a crosstalk between the Dicer-1 and Dicer-2 pathways [10; 16] (Figure 1A). Furthermore, the authors show that loqs plays a critical role in the maintenance of germ-line stem cells. While the two Loqs isoforms interacting with Dicer-1 are present throughout the adult fly, they show sex-specific expression in the gonads. Mutant female flies are sterile and lack germ-line stem cells. Mutations in piwi, a member of the Argonaute protein family, result in a similar phenotype [17], implying that dicer-1 and loqs might function together with piwi.With the discovery of loqs, it now seems a general feature of RNase III enzymes to cooperate with dsRBD-proteins in RNA silencing. This raises a couple of exciting questions. Both groups highlight the homology of Loqs with the human dsRBD protein HIV TAR RNA binding protein (TRBP), which binds to a hairpin-shaped TAR RNA, implicated in the response to HIV infection [18]. Moreover, TAR RNA has recently been suspected to be a viral pre-miRNA [19]. The homology between Loqs and TRBP is especially intriguing, as there is no clear human homolog for R2D2. Furthermore, human Dicer is more closely related to Drosophila’s Dicer-1. As Dicer-1 and Loqs are required for miRNA- and siRNA-mediated gene silencing, and Dicer-1 even shows weak siRNA-generating activity in the absence of Loqs, it seems plausible that human Dicer could function in a fashion similar to Dicer-1. Dicer-2 may play a unique role in Drosophila. Thus, findings in Drosophila siRISC biogenesis should be applied carefully to the human system.Beyond that, Loqs may be involved in processing other types of non-coding RNAs. Further studies on other binding partners will certainly help to characterize its function in greater detail. There are more than 70 miRNAs known in Drosophila to date [20], of which all pre-miRNAs fold into precursor hairpins which do not share common sequence motifs. So how does the Dicer-1-Loqs complex specifically recognize its substrates? By now, this question cannot be answered. Analyses of the double-stranded stem as well as the terminal loop structure of pre-miRNAs will make it possible to elucidate the precise molecular mechanism of substrate recognition. | |||||||
| PLoS Biol, 3(7): e244 (2005) [PDF]SynopsismiRNA Processing: Dicer-1 Meets its Match.DOI: 10.1371/journal.pbio.0030244In recent years, the control of gene expression by small RNA molecules has emerged as a major new mechanism for gene regulation. The small RNAs interfere with the expression of their target gene by reducing its transcription, triggering the destruction of the gene transcript, or inhibiting its translation into a protein. This discovery has not only altered views of gene regulation, but also provided molecular geneticists with powerful new tools with which to study and manipulate the function of any gene. The biology of these small RNAs is, therefore, under intense scrutiny.Small RNAs are generated by specific pathways, the elements of which are being rapidly discovered. In this issue of PLoS Biology, two groups have identified a missing piece in one such pathway- in the fruitfly Drosophila. The pathway under investigation leads to the production of a type of small RNA called a microRNA (miRNA). These are 21-23 nucleotides in length, and are involved in regulating the expression of many genes. miRNAs start life as a much bigger transcript called a pri-miRNA, which is processed in two steps. First, it is converted into a shorter pre-miRNA, by the action of two proteins: Drosha, an RNAse III enzyme; and Pasha, which contains double-stranded RNA binding domains (dsRBDs). The pre-miRNA is then transported to the cytoplasm and is trimmed again into a double-stranded miRNA by a different RNAse III enzyme called Dicer-1.In a separate pathway, RNAs called small interfering RNAs (siRNAs) depend on the Dicer-2 RNAse III and a dsRBD protein called R2D2 for their function. These pathways are also conserved in other organisms. Thus, a pattern emerges: the functions of small RNAs tend to require the combined actions of an RNAse III and a dsRBD protein. But why doesn’t Dicer-1 have a partner? The answer, provided by the two studies from the labs of Phil Zamore and Haruhiko and Mikiko Siomi, is that we just hadnケt found it yet.
The two groups took different approaches to finding Dicer-1’s partner. Zamore’s group looked for genes resembling other dsRBD-encoding genes, while the Siomi lab did a functional screen for new genes specifically implicated in miRNA processing. They both homed in on a new gene with great similarity to R2D2, and showed that loss of function of the gene results in the accumulation of pre-miRNAs-very similar to loss of Dicer-1 function, which suggests that the two genes act together in the same pathway. The new potential partner of Dicer-1 was given the name loquacious (loqs), because failure to process the miRNAs in turn causes increased levels of expression of the target genes for the miRNAs. Both groups also show that Loqs and Dicer-1 exist in a complex within the cell, and that the complex is able to process pre-miRNA into its mature form. The Siomi’s lab went on to show that the complex contains a protein called Ago-1, which hints that the complex might also be involved in the action of miRNAs on their target genes, as well as in miRNA processing itself. Both groups also point out the similarity between Loqs and a human dsRBD protein called TRBP, which has been implicated in the response to infection by HIV. There seems little doubt, then, that Dicer-1’s partner has been found, and that the combined action of an RNAse III and a dsRBD protein is a consistent theme in the function of miRNAs and siRNAs. The identification of Loqs will help to refine our views of how miRNAs are processed, as well as how they can be manipulated. The connections made with processes such as stem cell maintenance (identified by the Zamore lab) and viral infection in these new studies also emphasize that gene regulation by small RNAs is relevant to a broad range of cellular physiology. Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, et al. (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. DOI: 10.1371/journal.pbio.0030236 Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of premicroRNAs by the Dicer-1-Loquacious complex in Drosophila cells. DOI: 10.1371/journal.pbio.0030235 |
|||
| The Journal of Cell Biology, Volume 166, Number 3, 308-309 [PDF]Meeting Report West Meets East : JSCB 2004 Osaka, Japan May 26-28, 2004 Silencing specificity |
||||||
| RNA-mediated silencing begins with two types of RNAs. Those completely complementary to their target (siRNAs) lead to message degradation, whereas those less perfectly matched (miRNAs) block translation of their target. Both RNAs are produced by and carry out their very different functions within an RNAi-induced silencing complex (RISC). Until recently, all RISCs were considered equal. But evidence presented by Mikiko Siomi (University of Tokushima, Japan) suggests that RISCs are customized for the two RNAs with an appropriate Argonaute (AGO) family member.Flies have multiple AGOs, but Siomi shows that at least AGO1 and AGO2 are not interchangeable. AGO2 mutants were blocked in single-stranded siRNA production but could make miRNAs. The siRNA duplex is normally made by Dicer-2 cleavage of a long dsRNA, and AGO2 was needed to unwind this siRNA duplex, although no helicase domains have been identified in AGO2. |  |
|||||
| AGO1, in contrast, was needed for miRNA accumulation and formation of the miRNA-containing RISC, but was dispensible for siRNA function. AGO1 associated with Dicer-1, which cleaves the miRNA precursor, and with both unprocessed and mature forms of the miRNA. In the AGO1 mutant, there is much less mature miRNA, but its precursor did not accumulate in its place, so Siomi supposes that AGO1 stabilizes the mature miRNA after cleavage.The separate AGO functions suggest that perhaps one RISC does not fit all. “I believe that the [RISCs] are distinguishable in terms of the protein components,” says Siomi, “because we do not detect AGO2 in the AGO1 complex, and vice versa.”Making the RNAs is probably restricted to the complex with the appropriate AGO. The two complexes may talk to each other during later stages of silencing, however. “Perhaps at the second round or later … , the small RNAs are somehow exchangeable between RISCs, since we can clearly detect mature miRNAs both in the AGO1- and AGO2-containing complexes,” says Siomi. “But this is merely our speculation.” The biological significance of any such interchange remains to be determined.Reference:Okamura, K., et al. 2004. Genes Dev. doi:10.1101/gad.1210204. | ||||||
| Nature Reviews Genetics 5, 638-639 (2004) [PDF]RNA WORLDRNA stories on a mythical scaleMagdalena SkipperRNA interference (RNAi) hardly disappears off the science headlines. Most recently, the spotlight has been on Argonaute proteins ? some of which are involved in the small interfering RNA (siRNA) and microRNA (miRNA) pathways. Okamura and colleagues reveal an interesting division of labour between the Argonautes in Drosophila melanogaster: Argonaute1 (AGO1) is required for miRNA maturation and miRNA-directed RNA cleavage, whereas Argonaute 2 (AGO2) acts in the siRNA pathway.siRNA… miRNA… the difference is more than semantic. siRNAs are the key agents for RNAi and mediate RNA destruction in a sequence-specific manner. Although miRNAs can direct RNA cleavage, they can also block translation of their targets. Both species of small RNAs carry out their functions as part of the RNA-silencing complex (RISC), a multi-protein complex that mediates RNA cleavage. Given that both types of small RNAs associate with the same RISC, do the two pathways converge at this level, or do they differ?Prompted by previous studies in the worm, Okamura and colleagues decided to resolve the issue by looking at Ago1 and Ago2 mutants in Drosophila. They mobilized P-elements to delete Ago2 and found that it is required for RNAi in vivo and for RISC assembly; in particular, AGO2 is required for the unwinding of the siRNA, which is a prerequisite for siRNA-mediated cleavage. Although the RISC is associated with both siRNAs and miRNAs, Ago2 is required exclusively for the siRNA pathway.AGO1, on the other hand, seems to be involved only in the miRNA pathway. Ago1 mutant flies die as larvae with many developmental defects, and miRNA cleavage does not occur in lysates from Ago1 mutant embryos. In vivo analysis indicates that AGO1 directly interacts with Dicer-1, which is required for miRNA production from larger precursors, and that the role of Ago1 is to stabilize mature miRNAs. | ||||
| In mammals, siRNA and miRNA pathways seem to converge downstream of Dicer. This is not the case in other organisms, such as worms and flies. Okamura and colleagues have elegantly shown that it is Ago1 and 2 (as part of RISC) at the heart of the difference between siRNA- and a miRNA-mediated RNA cleavage. Undoubtedly, the small RNA world has many more equally elegant stories to tell us.ORIGINAL RESEARCH PAPEROkamura, K. et al. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655-1666 (2004) |  |
|||
| Modern Drug Discovery 72 May 2003 [PDF]Diseases and disorders: Fragile X syndrome.Randall C. Willis.Fragile X syndrome (FXS) is the most common form of inheritedmental retardation. An X-chromosome-linked disease, FXS is moreprevalent and its symptoms are more severe (see box, “Symptomsof fragile X syndrome”) in the male population (1:2000) than in thefemale population (1:4000). In 1991, Stephen Warren and colleagues at the Howard Hughes Medical Institute (Chevy Chase, MD) identified mutations in a gene (FMR1) located on the X chromosome of individuals with the disease. Unlike most disease-associated mutations, however, the FXS mutation resides in the gene-expression control region and takes the form of highly repetitive stretches of the nucleotide triplet CGG (Figure 1). Normally, this triplet repeats 5-54 times, whereas in “premutation” individuals, this sequence repeats 50-200 times. In FXS-affected or “full mutation” individuals, however, the repeat occurs more than 200 times.In the case of FMR1, the extreme number of CGG repeats causes excessive cytosine methylation in the promotor region of the gene such that FMR1 is not transcribed at sufficient levels. The lack of FMR1 mRNA leads to an equivalent or more severe lack of the protein encoded by the geneナ\the fragile X mental retardation 1 protein, or FMRP. FMRP is found predominantly in the cellular cytoplasm but can also translocate to the nucleus. Several research groups have shown that FMRP can bind to mRNA, but it preferentially binds to mRNA isolated from brain tissue.In 1999, Warren’s group used antibodies to isolate other mRNAbound proteins that might interact with FMRP. Among the proteins associated with FMRP, the researchers found two that belong to the same family of proteins as FMRP, as well as nucleolin and a protein involved in mRNA unwinding during translation (Figure 2). This indicated a possible role for FMRP in translation control.In early 2002, Peter Carlen and colleagues at the Toronto Western Hospital (Ontario) linked FMR1 in mice to the function of the GluR1 protein, a neurotransmitter involved in learning. The researchers found reduced levels of GluR1 protein in brain tissue from mice that lacked FMR1, which resulted in an inability of the tissue to potentiate applied electrical signals-an in vitro model for learning. Other studies have similarly located FMRP at the neuronal synapse, and its deletion has been shown to alter synaptic plasticity, which is associated with learning and memory.Also in 2002, Haruhiko Siomi and colleagues at the University of Tokushima (Japan) purified a dFMR1 complex from Drosophila cell lysates that contained two ribosomal proteins. But they also found a third protein that is involved in RNA interference (RNAi), a cellular mechanism that inhibits infections and might be involved in mRNA degradation and therefore post-transcriptional gene control. Although the fly homologue of FMRP did not seem to be essential for RNAi-mediated mRNA degradation, it might play a secondary role. This finding might have serendipitously led researchers to the real cause of FXS, a defect in RNAi activity in neurons.
Unfortunately for people with FXS, there are still as many questions as there are answers. The complexity of the symptoms and the unknown pathogenesis of the syndrome complicate FXS treatment, because a palliative effect on one symptom can often worsen other symptoms. That being said, stimulants, selective serotonin reuptake inhibitors, anticonvulsants, and antipsychotics have all been used to attack some of the symptoms associated with FXS. But as concluded at a National Institute of Mental Health (Bethesda, MD) conference on FXS, “There is [still] a critical need for controlled studies of pharmacological and behavioral treatment approaches for the behavioral and psychiatric manifestations of FXS.” |
|||
| Current Biology, Vol 12, R852-R854, 23 December 2002 [PDF]DispatchRNA Interference: The Fragile X Syndrome ConnectionRichard W. CarthewFragile X syndrome is caused by loss of expression of FMRP, a protein proposed to act as a regulator of mRNA translation which promotes synaptic maturation and function. Now FMRP has been found to associate with the RNP complex that mediates post-transcriptional silencing by RNAi. | |||
| science, Vol 298, Issue 5593, 497 , 18 October 2002 [PDF]Highlights of the recent literatureEDITORS’ CHOICEFragile X syndrome is the most common form of inherited mental retardation and is generally caused by mutations in the fragile X mental retardation gene (FMR1). The FMR1 protein is thought to negatively regulate the translation of specific genes. Caudy et al. and Ishizuka et al. have now demonstrated that the homologous protein in Drosophila, dFMRP, is associated with a vital component of the RNA interference (RNAi) machinery, the RNA-induced silencing complex (RISC), as well as with ribosomal proteins and VIG (Vasa intronic gene). These results suggest that RISC might provide a link between RNAi-based and FMRP-based modes of post-transcriptional gene control or, more provocatively, that FMRP may play a role in RNAi, thereby implicating defects in RNAi in human disease. — GRGenes Dev. 16, 2491; 2497 (2002) | |||